Assessing Microbial Colony Counting: A Deep Learning Approach with the AGAR Image Dataset
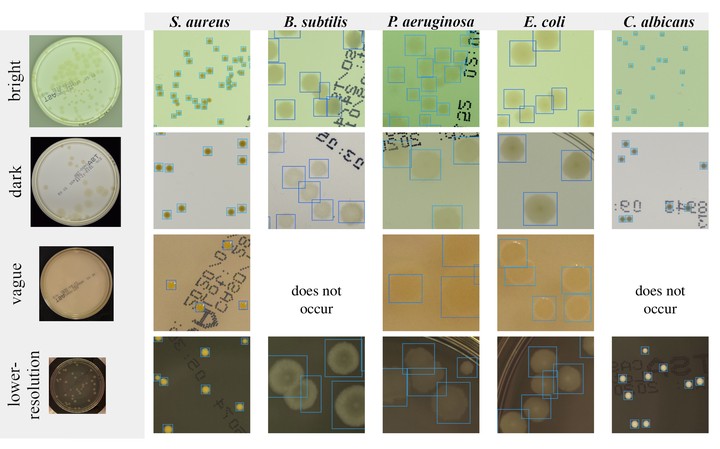 Image credit: Neurocomputing
Image credit: NeurocomputingAbstract
This study introduces the Annotated Germs for Automated Recognition (AGAR) dataset, an image database designed to advance the automation of microbial colony detection and classification. It encompasses an extensive collection of 18,000 high-resolution images depicting five distinct microorganisms, meticulously curated as single or mixed cultures, taken under diverse lighting conditions using two different camera setups. The dataset categorizes images into countable, uncountable, and empty, with the countable images further labeled by expert microbiologists to mark colony locations and species identification, totaling 336,442 colonies. We detail the comprehensive methodology employed in the creation of the AGAR database, emphasizing the rigorous annotation process by a team of professional microbiologists who have worked with microbial collection and identification for over 20 years. This process ensures the high-quality data necessary for training machine learning models. Our investigation further explores the performance of deep neural network architectures for object detection, namely Faster R-CNN and Cascade R-CNN with four different backbones, in leveraging the AGAR dataset for the tasks of microbial colony localization and species classification. The results confirmed the great potential of deep learning methods to automate the process of microbe localization and classification based on Petri dish photos. AGAR stands as the first dataset of this kind, both in scale and in public availability, laying the groundwork for future advancements in the field of machine learning applied to microbiology.