Recognition and identification of bacterial colonies
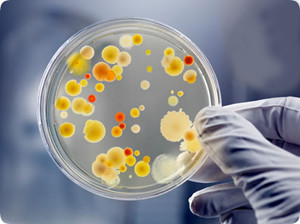 Photo puzzlefry.com
Photo puzzlefry.comRecognition and identification of bacterial colonies
Counting bacterial colonies is a fundamental task in microbiology, which is currently performed manually in most laboratories. This is a time-consuming and error-prone process, which requires a trained professional.
As one of our internal R&D projects, partially funded by The National Centre for Research and Development, we work on a library for automatic identification and classification of bacterial colonies based on RGB images of Petri dishes. Our deep learning methods allow us to detect and count different types of microorganisms with high accuracy and can be easily integrated with lab automation software or used as a standalone application.
What and why?
The Annotated Germs for Automated Recognition (AGAR) dataset is an image database of microbial colonies cultured on an agar plate. It contains 18000 photos of five different microorganisms, taken under diverse lighting conditions with two different cameras.
All images are classified into countable, uncountable, and empty, with the former being labeled by microbiologists with colony location and species identification (336 442 colonies in total). This study describes the dataset itself and the process of its development. Furthermore, the performance of selected deep neural network architectures for object detection, namely Faster R-CNN and Cascade R-CNN, have been tested on the AGAR dataset as well.
The results proved the great potential of deep learning methods to automate the process of microbes localization and classification based on Petri dish photos. Moreover, the AGAR is the first publicly available dataset of this kind and size serving for future development of machine learning models.
My contribiution
The aim of the project is to develop a method to automate the analysis of bacterial colonies on Petri dishes using artificial neural networks and Machine Learning algorithms, in particular, to classify the type of bacteria and to determine the number of colonies. I tested multiple detectors (also using cross domain detection and GAN/cycleGAN/bicycleGAN models), approaches based on density map estimation and segmentation.
Main tasks:
- Preprocessing photos to prepare specific annotations format for detection and instance segmentation
- Bacterial colonies localization and counting using fully supervised object detection and cross-domain object detection using unlabeled data from target domain
- Bacterial colonies counting by estimating a density map with convolutional neural networks
- Analyze point clouds data - classical machine learning clustering methods, like DEBSCAN
- Verifying possibility of using domain adaptation methods for training object detector in case of detection made on photos in different style (different illumination condition)
- Exporting the detector model via TorchScript to a production environment
- Preparing poster for ML in PL conference, and scientific paper
- Creating knowledge base
Technologies used: Python, Pytorch, OpenCV
Deliverables
Main works:
- Self-Normalized Density Map (SNDM) for Counting Microbiological Objects (2022)
- Generation of microbial colonies dataset with deep learning style transfer (2022)
- Deep neural networks approach to microbial colony detection – a comparative analysis (2021)
- AGAR a microbial colony dataset for deep-learning detection (2021)
- Counting and classification of bacterial colonies using deep learning (Conference MLinPL 2019)
Github code:
- Microbial dataset generation with style transfer
- Microbial counting review
- Objects counting by estimating a density map with CNNs
Medium posts:
- Recognition and Counting of Microorganisms on Petri Dishes | by Jaroslaw Pawloski and Sylwia Majchrowska | Towards Data Science
- AGAR — Annotated Germs for Automated Recognition | by Jaroslaw Pawloski and Sylwia Majchrowska | Towards Data Science
Learn more about the project
Project’s website: agar.neurosys.com